Projects

Detection of malarial parasites: We are focusing on developing a system to measure the degree of infection by malaria parasites using a scan of a colour photograph of stained malarial blood taken using a microscope in order to evaluate the parasitaemia of the blood, i.e. to count the number of parasites per number of red blood cells. A manual analysis of slides is tiring, time-consuming and requires expert technical staff. Our task is thus to automate the counting processes. Therefore, we want to count the red blood cells in addition to counting the parasites. Location is also important especially locating the parasites for visualization purposes.
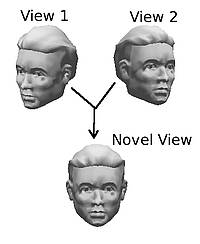
3D object recognition: Being able to recognise an arbitrary, real-world object is a fundamental endeavour of computer vision. However, this task requires the existence of a full 3d model of the object, which itself needs expensive and dedicated hardware. We are examining techniques, whereby we can combine a small number of images in such a way so as to generate valid, novel views of a 3d object and thereby recognise the object directly from its 2d images, without the need for a full 3d model.

Cell phenotype recognition: High throughput biological methods allow us to visualize the effect of knockdown of almost every gene in a genome via imaging fluorescently stained cells. However, such a dataset is so large that it is impractical for each picture to be analysed by a human expert. We aim to create a fast automated system to identify and characterize a phenotype based on its image properties.

Face recognition: Most face recognition algorithms perform very badly if the face they are supposed to recognize is not in exactly the same pose as the one in their database. The aim is to induce "pose invariance". In other words we want the computer to predict how the person will look from the side even if it only has one frontal snapshot.
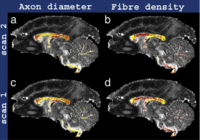
Microstructure imaging with diffusion MRI: Diffusion MRI is sensitive to the dispersion of diffusing water molecules within tissue. The tissue microstructure determines the dispersion pattern and thus the MR signal. We can solve an inverse problem to estimate and map histological features of tissue (cell size, shape, packing density, etc) from multiple diffusion MR images. The key application is non-invasive histology. Classical histology uses a microscope to reveal the cellular architecture of an excised tissue sample and provides the gold-standard diagnosis for a wide range of diseases, from cancers to dementias (such as Alzheimer's disease). Microstructure imaging potentially provides the same information non-invasively avoiding uncomfortable and risky biopsies. The image shows maps of axon density and diameter in the white matter of the brain. See the work of the Microstructure Imaging Group for more details.
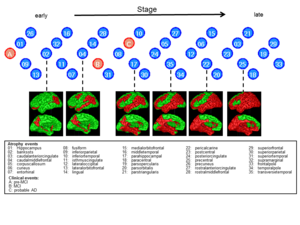
Disease progression modelling: The project aims to recover the series of events that take place as a disease plays out. In Alzheimer's disease for example, several symptoms and pathologies appear, such as memory impairment, personality changes, protein deposits in brain tissue, and regional grey matter atrophy. However, several other neurological diseases share these events. Their order of appearance and severity distinguish different diseases. However, current models of this progression are largely hypothetical. We use image analysis and machine learning to recover the series of events that characterise a particular disease from large cross-sectional data sets: typical data sets include many patients, but only acquired at a single time at some unique and unknown stage of the disease. The learning problem is to order the patients in terms of disease stage and then infer the characteristic progression pattern.
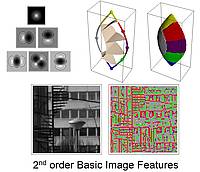
Basic image features: A fundamental 'alphabet' of image features is being derived based on geometric considerations and requirements of invariance. The feature alphabet is defined in terms of a gaussian derivative filter model of the operation of neurons in V1. The prefix 'basic' is applicable because of a deep and productive formal analogy between the feature categories of spatial vision and the basic colour categories (red, orange, etc.) of colour vision.
